The wobble hypothesis explains that the binding between the 3rd codon base and the 1st anticodon base does not follow canonical Watson-Crick base pairing, allowing multiple codons to code for a single amino acid.
There are 20 different amino acids that can make up a protein (22 if you count the rare amino acids: selenocysteine and pyrrolysine). Each amino acid is formulated with the help of ribosomes, mRNA and tRNA, which together make up the protein-generating machinery. Amino acids are added one after the other in the ribosome, like beads in a necklace. The resulting ‘necklace’ is a polypeptide chain that is then folded into a protein.
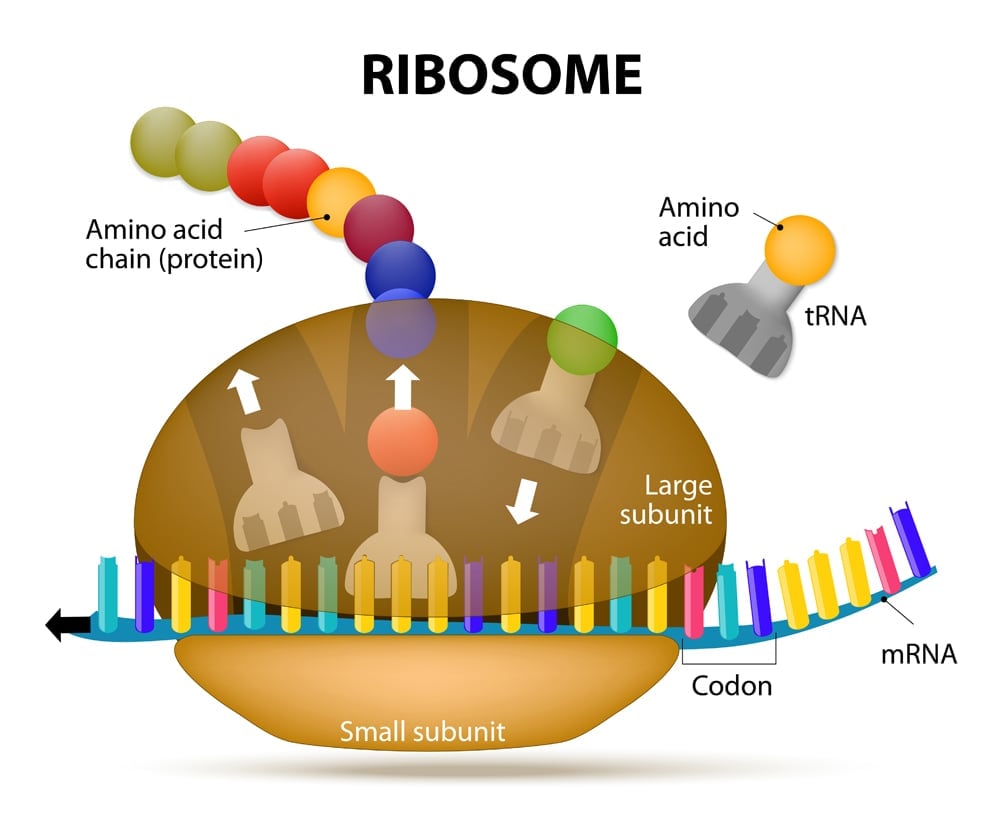
Before we uncover how multiple codons can code for a single amino acid, let’s learn a bit more about the players involved.
Recommended Video for you:
mRNA: The Starting Point Of Translation
Messenger RNA or mRNA is a single-stranded piece of RNA. It is synthesized during a process called transcription, where the information from the DNA is copied or transcribed into an mRNA strand. The double-stranded DNA helix opens up and an mRNA strand identical to the 5’ to 3’ coding strand is produced. Note that ribonucleic acid (RNA) contains the nucleotide uridine (U) in place of thymine (T), which is the only difference between the mRNA and the coding (5’ to 3’) strand of the DNA.
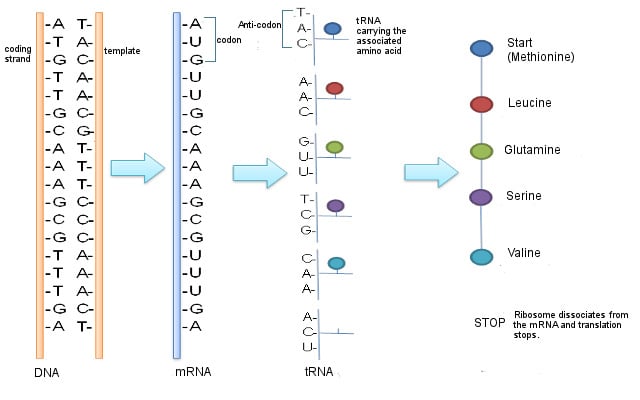
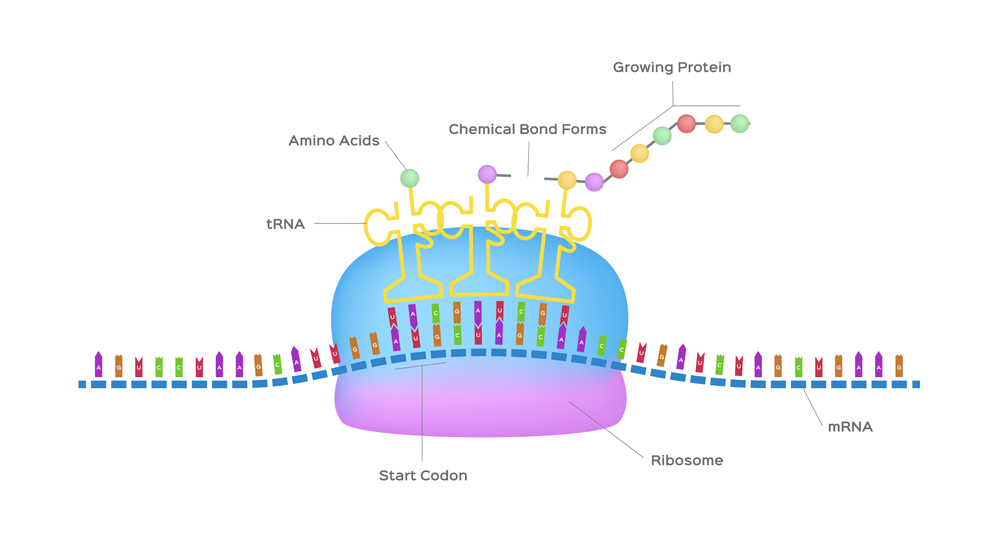
The mRNA then makes its way to the protein-making machinery—the ribosomes. The ribosomes read the information in mRNA to make proteins. This is called translation, like translating instructions to make something.
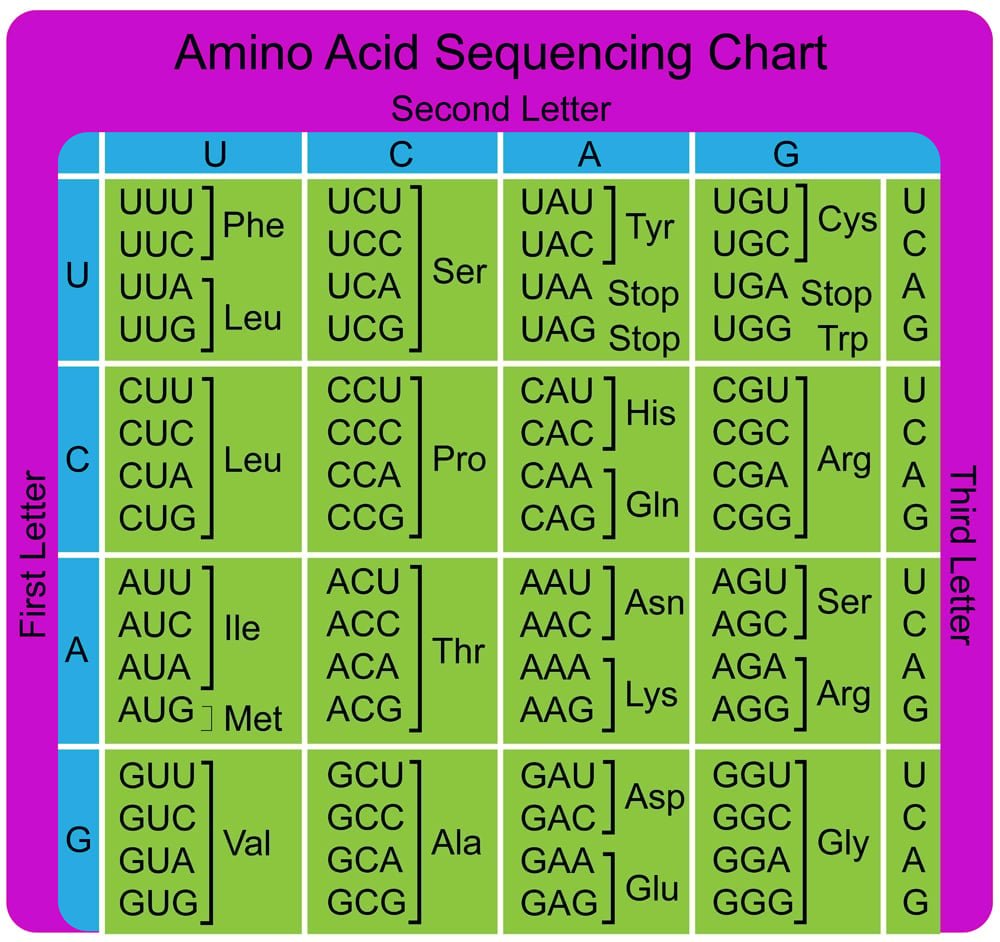
The information in the mRNA is read three nucleotides at a time in sections called codons. Each codon specifies a certain amino acid.
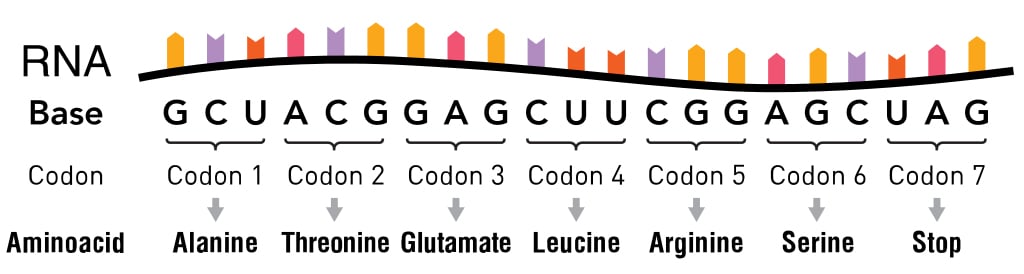
For example, the codon AUG codes for the amino acid methionine (Met). AUG also codes where the protein information starts, called the start codon, which is required to initiate the translation process. Thus, methionine is always the first amino acid in an amino acid chain. The amino acid tryptophan (Trp) is indicated by the codon UGG.
Codons don’t only code for amino acids; they also code for when to terminate the translation process. They are collectively known as stop codons and are UAG, UAA and UGA.
Hence, a codon is like a three-letter password that is required to obtain an amino acid and marks where the instructions to start and stop making the protein are found.
tRNA: The Assembly Point Of Translation
The transfer RNA or tRNA is also single-stranded, but is folded, unlike the straight mRNA. It serves as a physical link between the mRNA and the amino acids by interpreting the mRNA and transferring the right amino acids to their place while making the protein. If the translation machinery was a factory, the tRNA would be the factory workers who interpret the instruction manual (the mRNA) to carefully put products in the specified order.
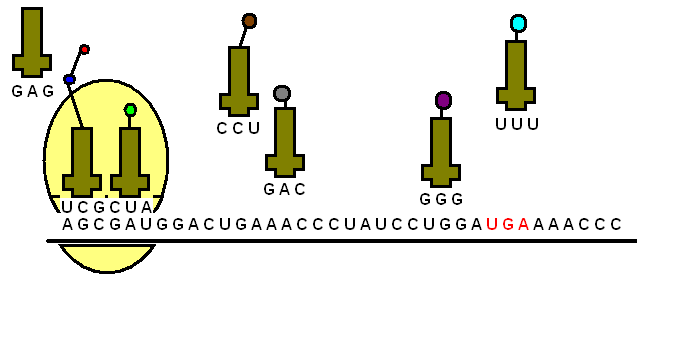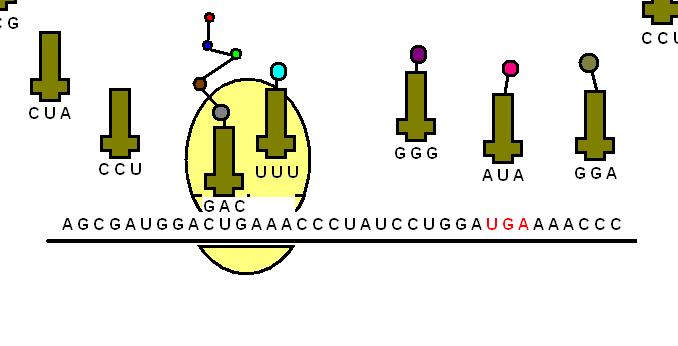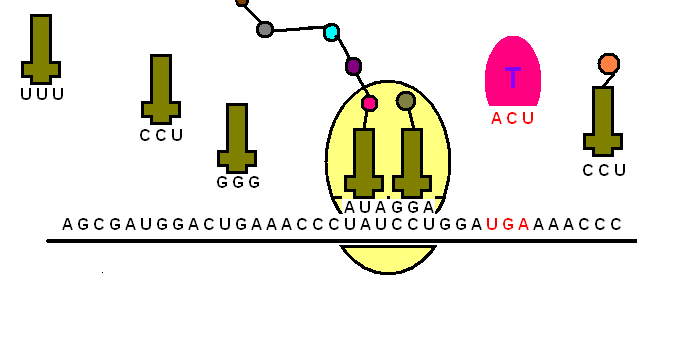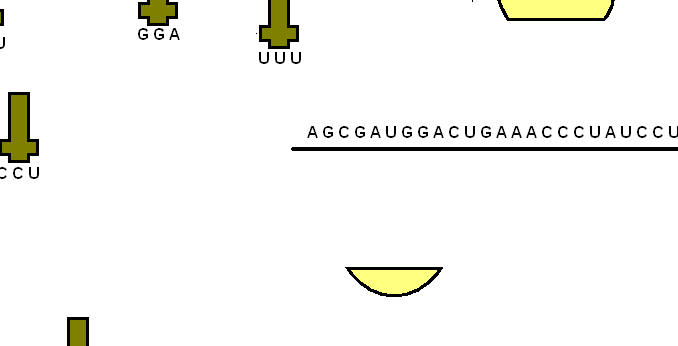
The tRNA is structured in such a way that it has an anticodon loop on one end and an acceptor arm/stem on the opposite end.
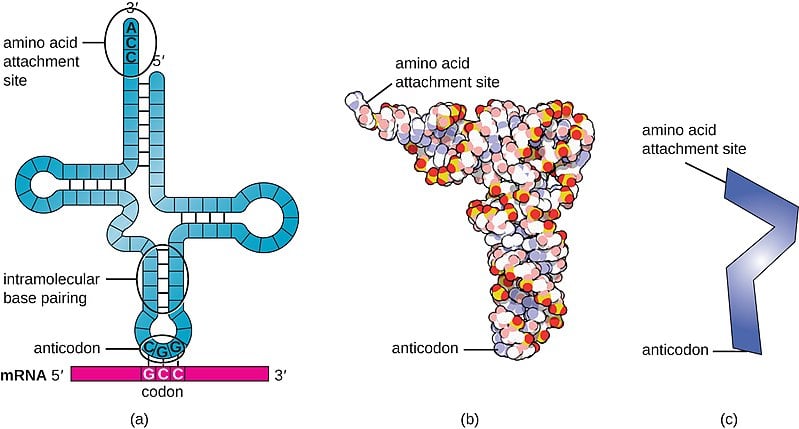
As the name suggests, the anticodon loop is complementary and antiparallel (3’ to 5’) to the mRNA codons. That is, the tRNA consists of the 3 nucleotides that bind to the ones present on the mRNA. Thus, guanine (G) and cytosine (C) bind to each other and adenine (A) and uridine (U) bind to one another.
So, considering the codon for methionine, AUG, it would have a methionine tRNA with an anti-codon UAG.
This type of pairing is known as Watson-Crick base pairing. The pairing is like the attraction between highly specific magnets. Anti-codons on the tRNA ensure that it lines up against the correct codon on the mRNA. Furthermore, as codons are read in the 5’ to 3’ direction, anticodons present on the tRNA are positioned in the 3’ to 5’ direction.
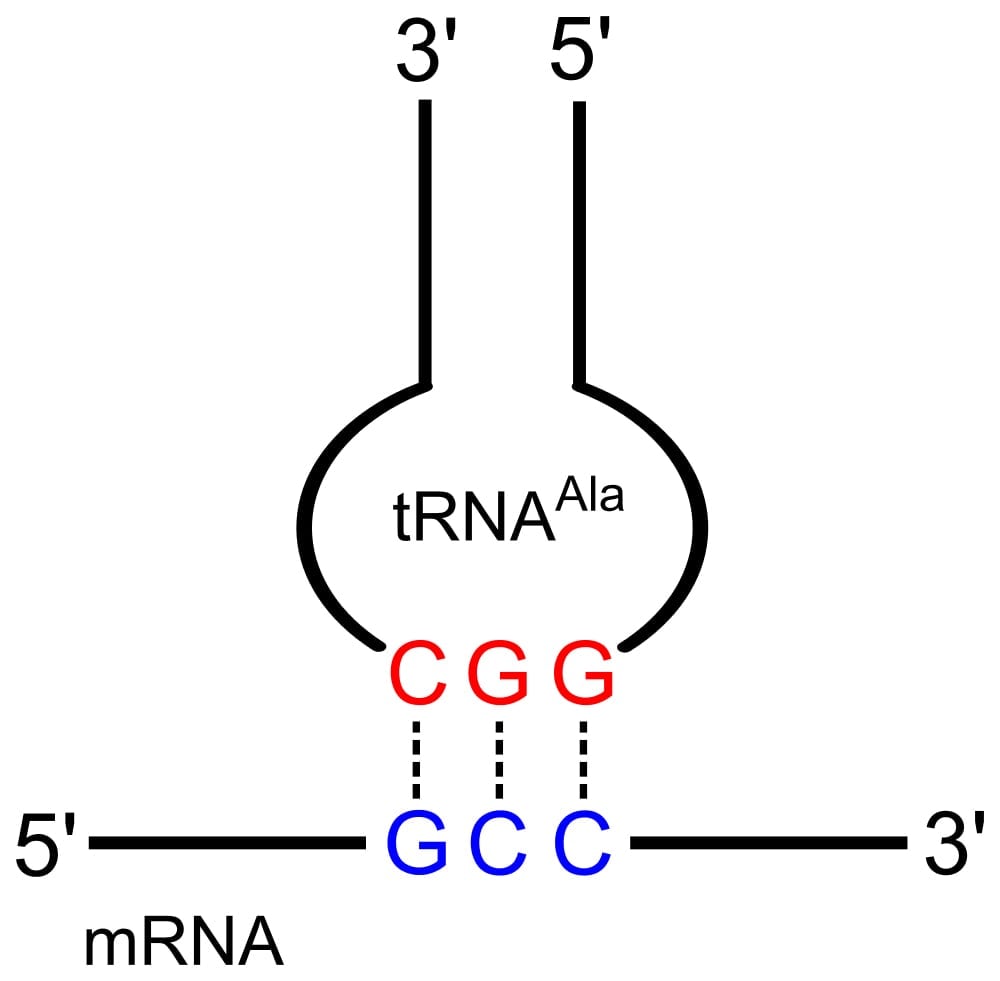
This means that the 1st codon base binds to the 3rd anticodon base and so on. Note that each amino acid has its very own tRNA, which correctly positions it in the polypeptide chain due to the base-pairing between the codon and anticodon.
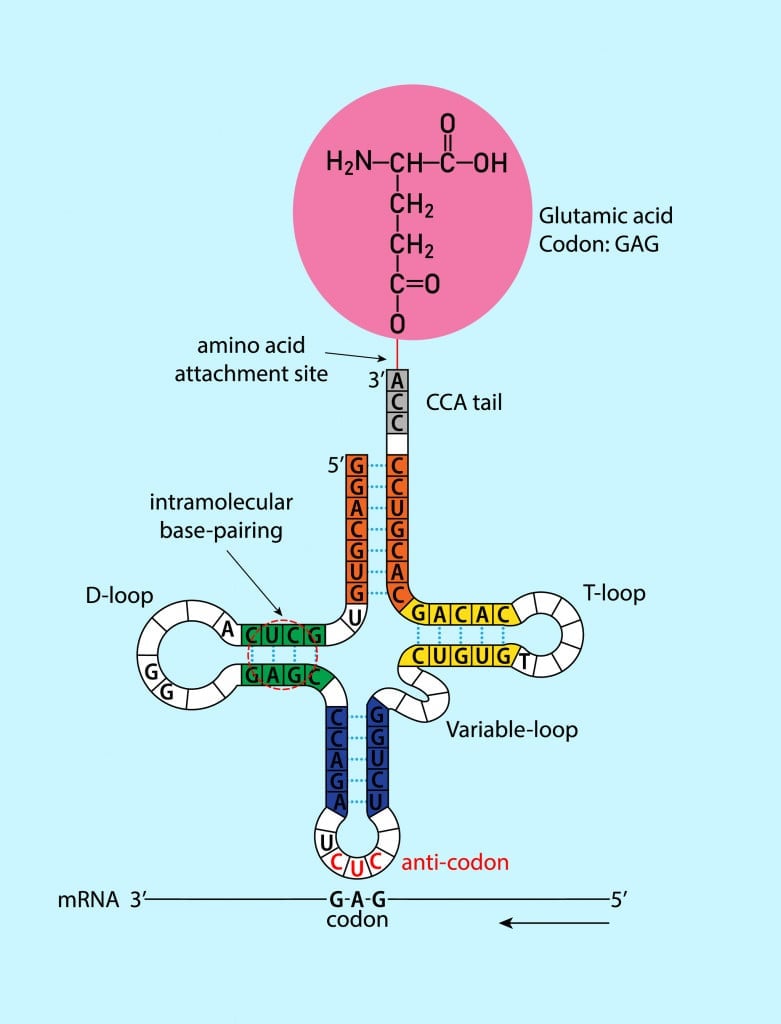
However, like many things in biology, this is also a little more complicated. Each amino acid can be specified by more than one codon. Additionally, the base-pairing rules between the codon and anticodon are not equally binding for all bases. We will learn more about this peculiar phenomenon next!
Wobble Hypothesis: Why 20 Amino Acids Have 64 Codons?
As we know, DNA is made up of 4 nucleotide bases (A, G, T, C). These letters (bases) are read three at a time, which means that there are 64 (4 x 4 x 4) possible combinations of these triplets or codons.
Of the 64 codons, 3 are stop codons, which we mentioned previously. These three stop codons do not code for amino acids and only terminate the process of translation. Therefore, we are left with 61 codons for just 20 amino acids.
The only logical option is that a single amino acid can be coded by multiple codons. But, in theory, 61 different tRNAs would be required for every different codon, yet this isn’t the case. Fewer than 50 codons have been identified in nature.
This observation implies that there might be some leeway between codons and anticodons matching up. Consider the amino acid valine, which is coded by 4 codons: GUU, GUC, GUA, GUG. Notice that only the third base changes, while the first two are the same.
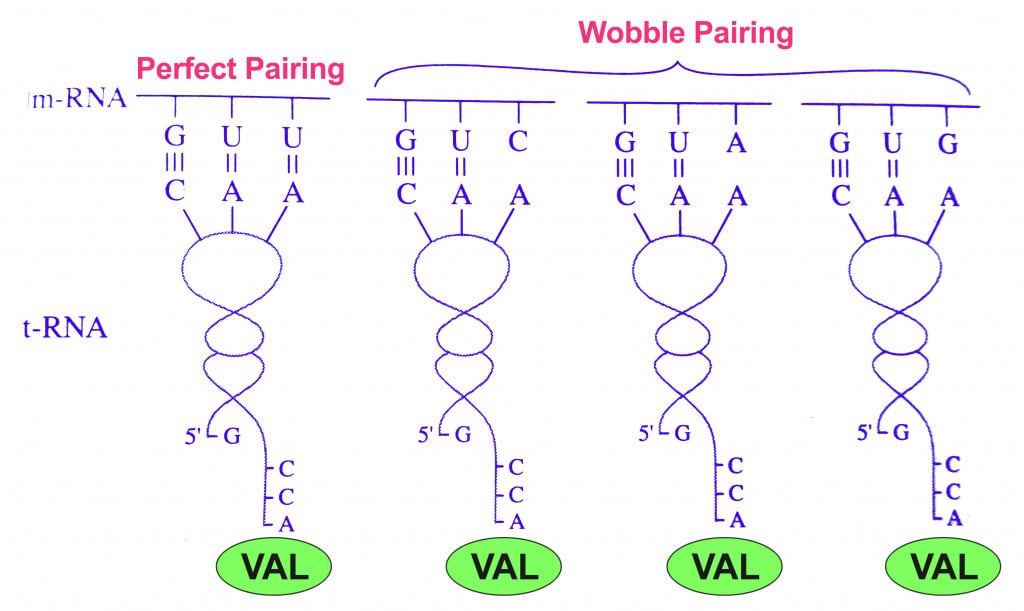
This variance observed only on the 3rd base of the codon led to the ‘Wobble Hypothesis’, proposed by Francis Crick (on of the discoverers of DNA structure) in 1966.
The Wobble Hypothesis explained this variance by revealing that the rules of Watson-Crick base-pairing were not followed at the last position of the codon. That is, the bond between the 3rd letter (nucleotide base) and the 1st letter of the anticodon may show unusual bonding (non A-T/ non G-C). Hence, it was allowed to ‘wobble’.
This meant that while the 1st and 2nd bases of the codons strictly adhered to the rules of Watson-Crick base pairing by binding to the 3rd and 2nd base of the anticodon, respectively, non-Watson-Crick base pairing was allowed between the 3rd base of the codon and 1st base of the anticodon. Hence, some tRNAs can recognize many different codons. It also explains the pattern of redundancy in the genetic code (many codons for a single amino acid).
The rules of the Wobble Hypothesis are as follows:
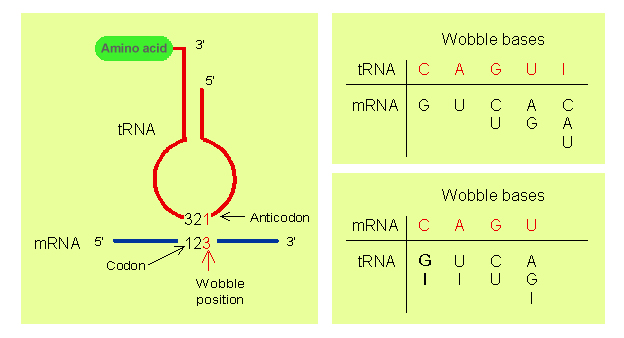
- The first two bases of the codon and the last two bases of the anticodon undergo normal Watson-Crick base pairing. That is, hydrogen bonds are formed between adenine (A) and uridine (U), guanine (G) and cytosine (C), only.
- Less stringent rules of base pairing apply at the remaining position and non-Watson-Crick base pairing may take place. The bases that undergo such pairing are also referred to as wobble base pairs. This allows the anticodon of a single form (amino acid specific) of tRNA to pair with more than one codon in the mRNA.
- When uridine (U) is present in the remaining (1st base of anticodon) position, it can recognize adenine (A) or guanine (G) only.
- When guanine (G) is present in the remaining (1st base of anticodon) position, it can recognize uridine (U) or cytosine (C) only.
- If the modified base inosine (I) is present in the remaining (1st base of anticodon) position, it can recognize uridine (U), cytosine (C) or adenine (A).
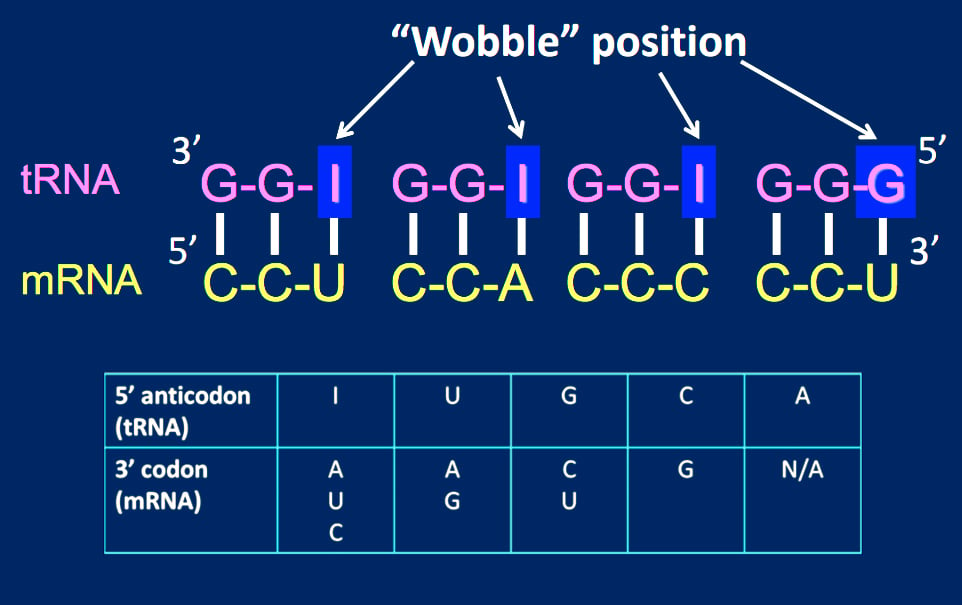
The reason for the wobble base pairing is a slip-up on the part of the ribosome. The ribosome has mechanisms in place to check whether the 1st and 2nd codon bases are complementary to the 3rd and 2nd anticodon bases, respectively. However, the ribosome doesn’t have a mechanism to check whether the 3rd codon base on the mRNA and 1st anticodon base on the tRNA match.
The Significance Of The Wobble Hypothesis
Slip-up or not, because of the wobble base, there is a lesser chance of mistakes during translation. For example, if the leucine (Leu) codon CUU was misread as CUA, CUG or CUC during transcription, the codon would still be translated as leucine (Leu) during protein synthesis.
Furthermore, protein synthesis is also safeguarded against mutations of a single base. Thus, if the 3rd base in the valine codon GUU mutates and changes to GUC, GUA or GUG, it will still be translated correctly.
Wobble and the degeneracy of the genetic code also reduce the number of tRNAs required by a cell. For instance, for the four different codons of glycine, there are only 3 tRNAs present in E.coli.
So, while it is true that multiple codons can code for a single amino acid, it is the ‘wobble’ observed between the 3rd codon base and the 1st anticodon base that makes this flexibility possible.
References (click to expand)
- Crick, F. H. C. (1966, August). Codon—anticodon pairing: The wobble hypothesis. Journal of Molecular Biology. Elsevier BV.
- (2014) Intronic and plasmid-derived regions contribute to the large .... National Center for Biotechnology Information
- BB331 Lecture 12 - oregonstate.edu
- How the ribosome decodes a modified 'wobble' position .... The European Synchrotron Radiation Facility
- Wobble Hypothesis - Newcastle University. Newcastle University
- Chapter 5. Genetic Code, Translation, Splicing - Biology. Kenyon College
- Amino Acids Are Encoded by Groups of Three Bases Starting from a Fixed Point - Biochemistry - NCBI Bookshelf - www.ncbi.nlm.nih.gov











